RNA editing has become a generic term for a wide array of post-transcriptional processes that change the mature RNA sequence relative to the corresponding encoding genomic DNA matrix. This phenomenon, which is almost limited to eukaryotes with some exceptions, is characterized by nucleotide insertion, deletion, or substitution in various types of RNAs including mRNAs, tRNAs, miRNAs, and rRNAs , and is likely to contribute to RNA diversity. Until recently, this mechanism was considered relatively rare in vertebrates, mainly restricted to brain-specific substrates and repetitive regions of the genome, and limited to extensively validated ADAR-mediated adenosine to inosine (A-to-I) substitutions and APOBEC-mediated cytosine to uracil (C-to-U) changes.
Since 2009, the advent of high-throughput sequencing technologies has enabled the study of this phenomenon at a transcriptome-wide scale and progressively challenged this view, with estimates ranging from several hundred to several thousand, and even millions of mRNA edited sites throughout mammalian genomes. According to some of these mRNA editing screening studies, mRNA recoding is an extremely common process that greatly contributes to transcript diversity. Furthermore, most of these studies report mRNA editing events leading to transversions that cannot be explained in the light of our current knowledge regarding the molecular bases of mRNA recoding, suggesting the existence of currently uncharacterized mRNA editing mechanisms and novel molecular components implied in gene expression regulation. The conclusions raised by these studies regarding the extent and nature of mRNA recoding, if further supported, would deeply impact our understanding of gene expression regulation and transcriptional modification.
Facing contradictory results regarding the extent of mRNA editing, a large number of studies and comments have pointed to the requirement for comprehensive and rigorous bioinformatics pipelines to limit technical artifacts in editome characterization. Working with short-read sequencing data for the detection of polymorphisms requires careful dealing with technical artifacts related to mapping on paralogous or repetitive regions, mapping errors at splice sites, or systematic and random sequencing errors1. This is especially the case when screening for mRNA editing events, since all of these artifacts are likely to generate artificial discrepancies between genomic DNA and mRNA further interpreted as edited sites. In this context, the huge variation regarding the extent of intratissue and intraspecies mRNA editing revealed in the literature could be in part due to the varying level of stringency of bioinformatics filters used to control these error prone artifacts, and whether biological replication is considered or not.
In the scope of this project, we developped a rigorous strategy summarized in Figure 1 to identify mRNA editing using both mRNA and genomic DNA high-throughput sequencing, taking into account sequencing and mapping artifacts, as well as biological replicates, to control the false positive rate. To strictly control multimapping, we looked for mRNA sequences spanning edited sites in unmapped genomic DNA sequences, allowing the consideration of potential errors and gaps in the reference assembly that still represent roughly 15% of the chicken genome.
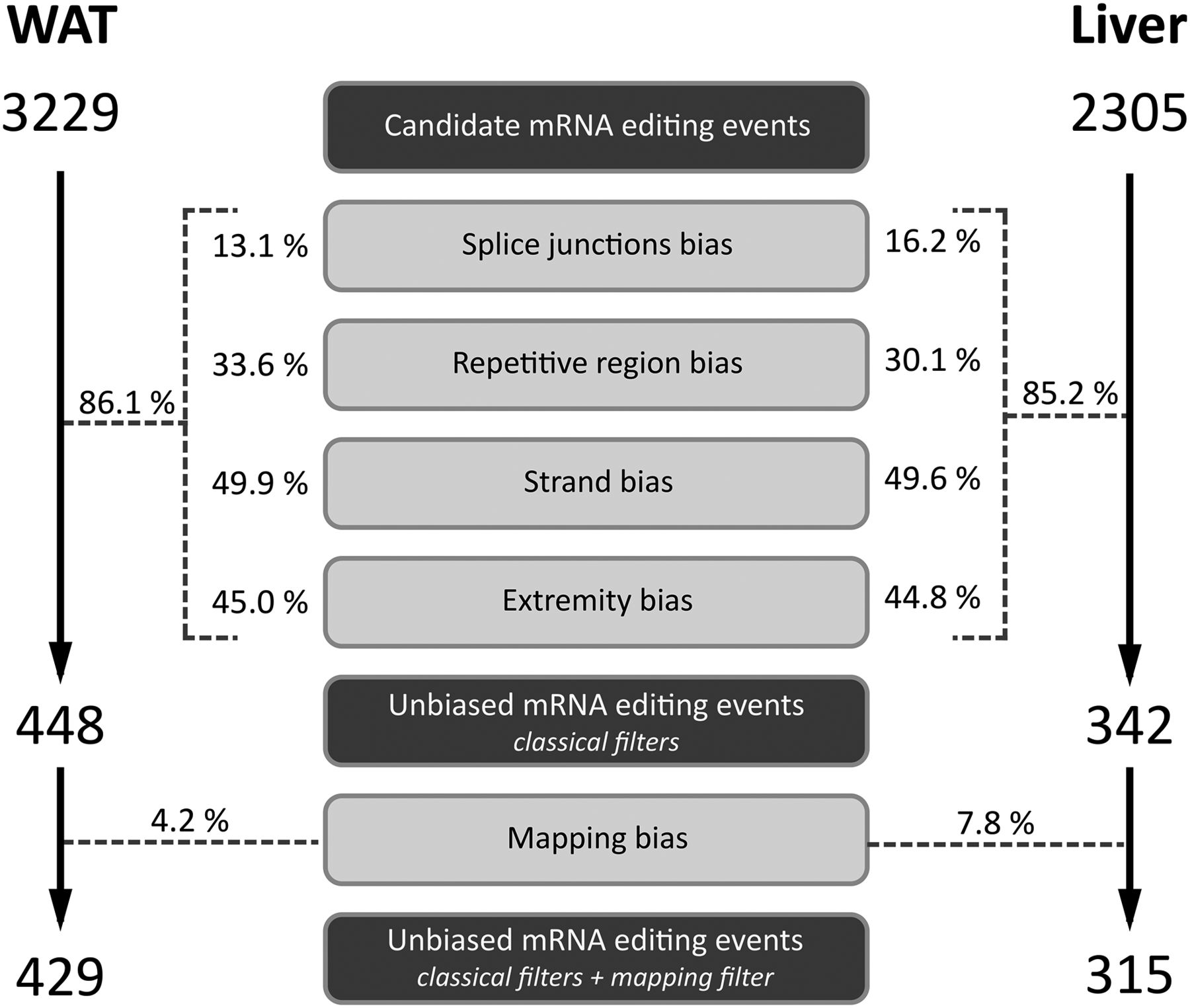
Impact of sequencing and mapping biases on mRNA editing discovery. Contribution of random or systematic sequencing biases and mapping artifacts to the false discovery of mRNA editing events using combined mRNA and DNA sequencings are given as a fraction (%) of the intial pool of candidate editing events subject to each source of bias in each tissue. WAT, white adipose tissue.
- For a comprehensive overview the scientific debate around the extend of mRNA editing, please refer to Schrider et al. 2011, Kleinman and Majewski 2012, Lin et al. 2012, Kleinman et al. 2012, Piskol et al. 2013, Lagarrigue et al. 2013 ^
